
Environment
Achieving environmental sustainability is one of civilization’s historic challenges. ISB uses systems science to improve the understanding of the interactions of microbes and ecosystems and create a new generation of sustainable tools and strategies that can be deployed to address this challenge.

Stress Test Predicts How Diatoms Will React to Ocean Acidification
The foundation of many marine food webs in which corals, fish and whales depend on are microscopic photosynthetic algae called diatoms. ISB scientists have developed a stress test to quantify and predict how ocean acidification would impact the resilience of diatoms in the future.
ISB Scientists Have Discovered When and Why a Microbial Community Might Collapse
Researchers at Institute for Systems Biology have developed a framework for assessing the “health” of a microbial community through a stress test that enables them to ask when and why microbial communities collapse under different environmental conditions.

Microalgae As Biofactories of a Sustainable Future
In an effort to better understand the gene regulatory and metabolic networks of the single-celled alga Chlamydomonas reinhardtii, researchers at Institute for Systems Biology studied the changes in Chlamy’s genetics and metabolism that cause them to capture and store carbon dioxide.

Project Feed 1010 – Sustainable Agriculture
ISB is actively working on Project Feed 1010, a demonstration project and laying the experimental, computational, cloud-computing and social networking infrastructure for a first-of-its-kind network of farmers, scientists and educators to crowdsource sustainable agriculture research and education.
Genetic Switch May Help Marine Microalgae Respond to Higher CO2 Levels
Rapid climate change, including ocean acidification caused by increasing carbon dioxide (CO2) levels, is predicted to affect the oceans, sea life, and the global carbon cycle.
Marine microalgae, including diatoms, are responsible for converting CO2 into oxygen and biologically usable carbon through photosynthesis. How these organisms will respond over the short and long term to rising CO2 is unknown.
Growth experiments and transcriptomic analyses performed by UW and ISB researchers showed that the model diatom Thalassiosira pseudonana respond to increasing CO2 by down-regulating gene clusters involved in carbon concentrating mechanisms (CCMs) and photorespiration. It is hypothesized that this acclimation may allow diatoms to conserve energy at moderately increased CO2 levels, with implications for shifts in the composition and productivity of marine microbial ecosystems.
Most Powerful Tool for Reconstructing a Gene Network
Nearly a decade ago, ISB’s Baliga Lab published a landmark paper describing cMonkey, an innovative method to accurately map gene networks within any organism from microbes to humans.
Two new papers describe the benchmark results of cMonkey and also the release of cMonkey2, which performs with higher accuracy.
Using this approach, genetic and molecular data generated from any organism, be it a bacterium or a birch tree, can be explored and analyzed from a network perspective.

A Universe Under the Microscope
It may seem strange to begin a prognostication for microbiology with a quote from an astronomer, but the two topics are more closely linked than appears at first glance. For starters, both fields were revolutionized around the same time, in the 17th century, with drastic improvements to the telescope and the compound microscope.
How One Family of Microbial Genes Rewires Itself for New Niches
When an organism duplicates its genes, it increases its ability to adapt and colonize new environments.
ISB researchers used the systems approach to study how one family of microbial genes evolved to bring functions that were adaptive to specific environments.
This new understanding of how gene regulatory networks rewire themselves has many potential applications, including how to wire new functions into an organism for biofuel production, bio-remediation or bio-pharmaceutical production.
New Tool Uses 3-D Protein-DNA Structures to Predict Locations of Genetic ‘On-Off’ Switches
Novel systems approach uses high-resolution structures of protein-DNA complexes to predict where transcription factors (genetic switches) bind and regulate the genome.
This approach can help researchers better understand and predict binding sites for non-model organisms or ‘exotic’ species.
Having such insight and predictive capabilities is critical for reverse- and forward-engineering organisms that could be pivotal for new green biotechnologies.

New Open-Access Multiscale Model Captures Dynamic Molecular Processes in Unprecedented Detail
Microbes are efficient because their streamlined genomes allow them to evolve and adapt rapidly to complex environmental changes.
Decoding the highly-compressed information within a microbial genome requires sophisticated systems biology tools to map the genetic programs, and understand how they are executed.
ISB researchers invented novel algorithms to unzip and decode microbial genomes into the EGRIN 2.0, an open-access multiscale model that captures instructions for executing the dynamic molecular processes in unprecedented detail.
Unpredictable Environments Require Predictably Fast Responses
ISB researchers discover a novel mechanism used by cells to rapidly turn “on” or “off” genes in order to change survival strategies in response to environmental events.
The “switch” was discovered in Halobacterium salinarium and found to be conserved in diverse life forms.
Research shows that microbes may use specialized switches depending on the type of environmental challenge they encounter.

50 Years in the Lab Doesn’t Diminish an Organism’s Memory of Its Natural Environment
ISB researchers discover that microbes retain complex survival traits despite having been domesticated for more than 50 years.
Study reveals that gene networks store a historic record of environmental change.
Research reinforces the concept that model organisms can serve as authentic systems to study evolution of complex traits.
New Predictive Model for Gene Regulation of Methanogenesis of M. maripaludis
Methanogens catalyze the critical, methane-producing step (called methanogenesis) in the anaerobic decomposition of organic matter. Scientists at ISB and University of Washington led a multi-institutional effort to generate the first predictive model for gene regulation of methanogenesis in a hydrogenotrophic methanogen, Methanococcus maripaludis. The research was published on Oct. 2, 2013, in the journal Genome Research. Most species of methanogens are hydrogenotrophic –meaning they use electrons from hydrogen gas (H2) to convert carbon dioxide to methane.
Some Algae Are Destined to Die for the Good of the Planet
Nearly 25 gigatons of carbon is cycled annually through the oceans, replenishing resources for a healthy planet. This process is carried out by interactions among different groups of microorganisms, each performing a different role in a network that has come to be known as the microbial loop. In a study published in the journal PLOS One, researchers at the Institute for Systems Biology report the discovery that programmed cell death of algae is potentially an important step in the microbial loop.
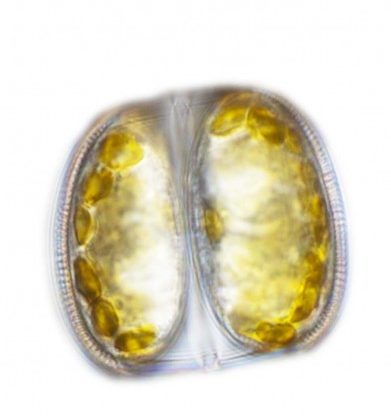
Discovery of Microbial Biological Clock May Shed Light on Ocean Health
Oceans contain innumerable species of diatom microbes that are responsible for half of the Earth’s oxygen production, as well as CO2 removal, a critical exchange in the web of life. The current understanding of these single-celled organisms is limited. But researchers at the Institute for Systems Biology (ISB) and the University of Washington have made a discovery that offers new insight about the daily life cycle of the diatom, which may lead to a better understanding of ocean health in response to ocean acidification and climate change, and potential applications for biofuel production.



