Baliga Lab: Uncovering the Genetic Adaptability of Tuberculosis
 isbscience.org/news/2014/09/18/baliga-lab-uncovering-the-genetic-adaptability-of-tuberculosis/
isbscience.org/news/2014/09/18/baliga-lab-uncovering-the-genetic-adaptability-of-tuberculosis/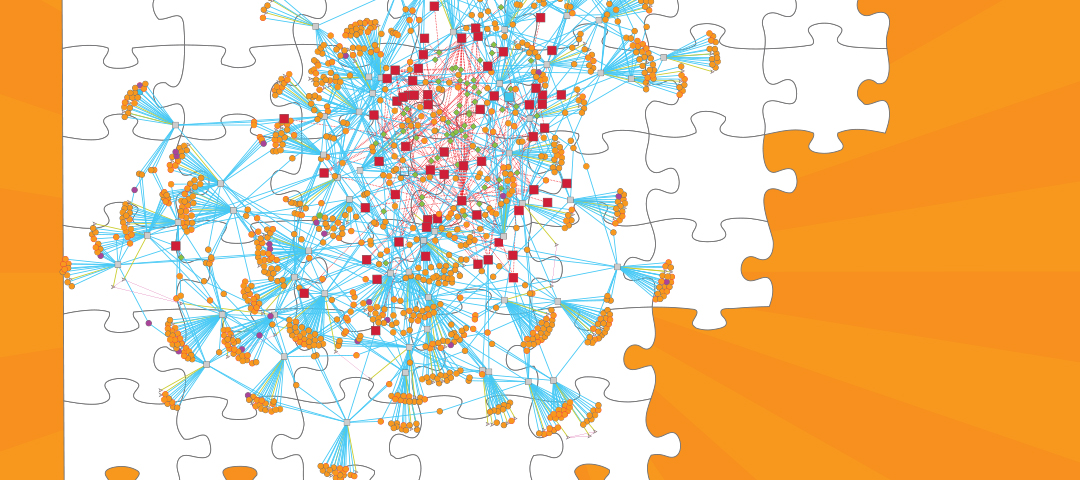
3 Bullets:
- The Institute for Systems Biology and Seattle BioMed have collaborated to reconstruct the gene regulatory network of the human pathogen Mycobacterium tuberculosis.
- Finely tuned gene regulation has allowed Mycobacterium tuberculosis to survive unnoticed in an apparently healthy host for decades; understanding those subtleties is critical for advancing treatment.
- The identification of co-regulated sets of genes and their regulatory influences offers validated predictions that will help guide future research into Mycobacterium tuberculosis pathogenicity.
By Jake Valenzuela and Eliza Peterson
Tuberculosis, responsible for nearly two million deaths each year, is caused by the bacterium Mycobacterium tuberculosis (MTB). Researchers from the Institute of Systems Biology in collaboration with Seattle BioMed have undertaken a comprehensive approach centered around the strengths of systems biology to identify adaptive regulatory mechanisms employed by MTB.
Journal: Nucleic Acids Research
Title: A high-resolution network model for global gene regulation in Mycobacterium tuberculosis
Authors: Eliza J.R. Peterson, David J. Reiss, Serdar Turkarslan, Kyle J. Minch, Tige Rustad, Christopher L. Plaisier, William J.R. Longabaugh, David R. Sherman, Nitin S. Baliga
Link: http://nar.oxfordjournals.org/content/early/2014/09/17/nar.gku777.full
The success of MTB as a human pathogen is largely due to its ability to alter gene expression and adapt to the complex host environment within which it resides. In an effort to model the gene regulation of this elusive microorganism, researchers compiled 2,325 publicly available gene expression profiles from a diverse set of conditions and constructed a gene regulatory network model of MTB. The gene regulatory network model predicts sets of genes that are similarly expressed, the proteins or factors that bind to DNA to influence their gene expression, and the environmental conditions in which these regulatory interactions occur. This global analysis gives us a clearer idea of MTB pathogenicity and, by engaging in a systems biology approach, we better understand how the pathogen responds to a diversity of stress conditions like low oxygen, host interactions, and nutrient availability. In particular, the researchers highlighted the importance of host cholesterol availability during infection and provided new details about the regulation of cholesterol utilization in MTB.
Importantly, the gene regulatory network of MTB was made publicly available (http://networks.isbscience.net/mtb/) to encourage exploration of the model. This will hopefully drive future efforts toward understanding how MTB responds to various environmental conditions, including those responsible for pathogenicity. This network also laid the foundation for future versions of the model that will incorporate new data and new modeling techniques.
Read more about ISB’s work with Tuberculosis: ISB Researchers Identify New Protein Modification Critical Growth of Tuberculosis Pathogen
About Dr. Jake Valenzuela and Dr. Eliza Peterson: Jake is a postdoc in the Baliga Lab and a member of the ISB Editorial Board. Eliza is postdoc in the Baliga Lab and lead author of the paper.






