Cancer Stratification: A Systems Approach
 isbscience.org/news/2014/05/12/cancer-stratification-a-systems-approach/
isbscience.org/news/2014/05/12/cancer-stratification-a-systems-approach/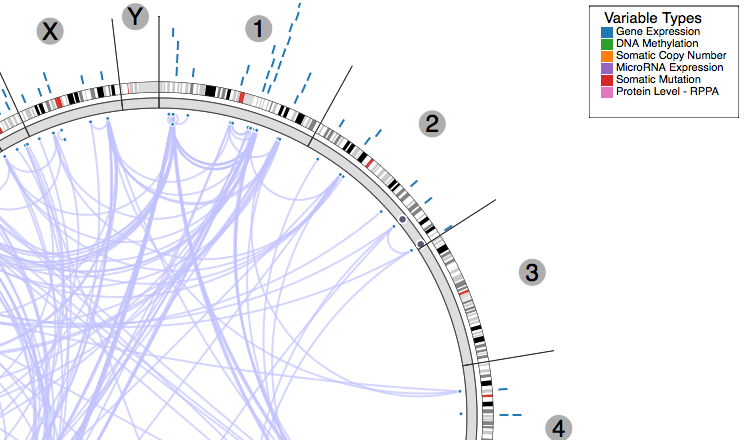
By Theo Knijnenburg
ISB Editorial Board Member
When a patient receives a diagnosis of breast cancer, it’s a specific subtype of breast cancer, such as invasive ductal carcinoma. Each subtype is characterized by the shape and location of the tumor, its growth progression, prognosis and treatment. The ability to stratify, or group, cancer patients based on the specific characteristics of their cancer type, is the first step toward personalized cancer treatment.
Whereas traditional stratification is based on histopathology — the examination of the tumor tissue — cancer medicine is rapidly moving toward stratification based on the molecular makeup of the tumor. A great example of the impact of molecular stratification is trastuzumab (Herceptin), an antibody that targets the extracellular domain of the HER2 receptor protein. Breast cancer patients with elevated levels of HER2 receive strong clinical benefits from treatment with trastuzumab. Novel targeted medicine, such as trastuzumab, will replace or complement chemotherapy, radiation therapy and surgery, the current standard-of-care in cancer treatment.
This is part of a series on ISB's systems biology approach to cancer research. Read the introduction or about
cancer detection and cancer treatment.
Stratification of cancer patients based on molecular data is, however, far from trivial. Not only does it require the careful collection and measurement of many thousands of cancer samples, but it further requires the use of novel and sophisticated computational approaches to identify relevant groups of samples as well as biomarkers that enable detection of these groups in the clinic. It’s necessary to integrate vast amounts of data and to be able to visualize that data in order to help stratify cancers.
To this end, researchers at ISB perform detailed analyses of individual tumors as well as global analyses of tumors from many different cancer types. On the scale of individual tumors, it is necessary to recognize that tumors contain multiple cell types, each with distinct aberrant genomic and/or proteomic profiles, and thus tumorigenic properties.
How ISB Applies A Systems Approach to Stratification
Tumor Heterogeneity
A central theme of the cancer and stem cell group at ISB led by Dr. Qiang Tian is to recognize the heterogeneity in a tumor and exploit this for improved molecular stratification of cancer. The team has demonstrated that isolated tumor-initiating cells with a single cell surface marker (e.g. CD133) can give rise to a gene expression signature, which enables better cancer grading and molecular subtyping (U.S. patent approved). Current efforts are focused on establishing single cell clonal cultures from primary tumor tissues and subjecting them to in-depth molecular profiling.
“…cancer medicine is rapidly moving toward stratification based on the molecular makeup of the tumor…”
The Cancer Genome Atlas
ISB is at the forefront of large scale analyses of tumor data. Specifically, the group of Ilya Shmulevich at ISB, in collaboration with the group of Wei Zhang at the MD Anderson Cancer Institute, is working as a data analysis center as part of The Cancer Genome Atlas (TCGA), currently the largest effort to comprehensively characterize the molecular landscapes of many different cancers. This characterization includes many different data types: Not only aberrations of the DNA in the tumor cells, such as mutations or amplification of large pieces of chromosomes, but also the expression of genes and the protein that the genes code for are analyzed. ISB is using sophisticated statistical pattern recognition and machine learning approaches on these data. This enables us to distinguish subtypes or different stages of a cancer, to determine whether a cancer has metastatic potential, and to predict survival or the likelihood of a successful response to a therapy.
Technology Innovation
ISB not only invests in data analyses, but also in making the data and results of the analyses directly and easily available to the community of cancer researchers at large. To that end, the ISB has developed several interactive visualization tools, such as Regulome Explorer and GeneSpot. Regulome Explorer is a cancer "omics" data browser – a web-based application that was created as part of the TCGA project to allow researchers to visually explore associations found among features such as gene (mRNA) expression, methylation events, microRNA expression, somatic mutations, copy number events, protein levels, and clinical parameters. GeneSpot provides a way to explore TCGA data from a gene-centric point of view, giving access to interactive visualizations for multiple data types and cancers. These tools provide insight into the large scale TCGA data and will help to stratify and ultimately treat cancers.
About Theo Knijnenburg: Theo is a postdoc in the Shmulevich Lab and is a member of the ISB Editorial Board.






