
Method Developed to Profile Pathogen Gene Expression from Infected Host Cells
The rise in multi-drug resistant and extremely drug resistant strains of Mycobacterium tuberculosis has become a major cause of global health concern for treating tuberculosis. ISB researchers developed Path-seq, a method that profiles host and pathogen gene expression and can explore the pathogen transcriptome in vivo.

Wilke Cohen Lyme Disease Project
ISB researchers have put their efforts into looking at Lyme disease and the bacteria, Borrelia burgdorferi, that causes the disease, through the lens of systems biology, and utilizing approaches pioneered at ISB to gather, synthesize and interpret large and complex datasets.

Identification of the proteins in Plasmodium vivax provide new targets for a malaria vaccine
Scientists from ISB and the Center for Infectious Disease Research led an international collaboration to identify proteins in the malaria parasite Plasmodium vivax. P. vivax and P. falciparum cause the majority of malaria cases, but P. vivax is far less-studied, in part because it cannot be grown in the lab. The research aims to provide new targets for a malaria vaccine.
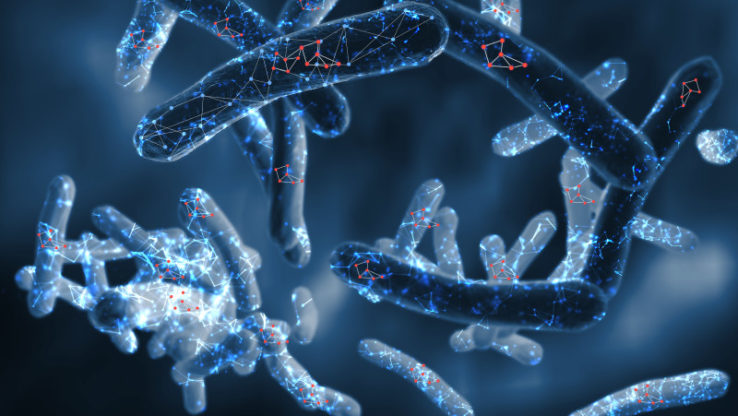
Speeding Up Drug Discovery to Fight Tuberculosis
Researchers at the Institute for Systems Biology and Center for Infectious Disease Research have deciphered how the human pathogen Mycobacterium tuberculosis is able to tolerate the recently approved FDA drug
The study demonstrated that silencing certain regulatory genes in the bacteria, or pairing with a second drug pretomanid, disrupts a tolerance gene network to improve efficacy of killing by bedaquiline.
This systems-approach to rational drug discovery represents significant advance in the fight against tuberculosis, which affects a third of the global population, surpassing HIV/AIDS in the number of deaths worldwide.
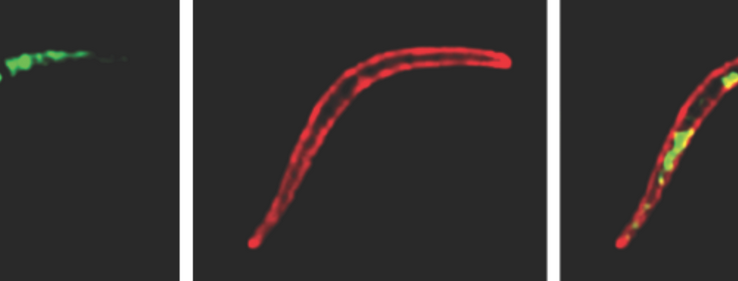
Scratching the Surface of the Malaria Parasite
Surface coat proteins on Plasmodium, the parasite that causes malaria, provide new targets for a malaria vaccine. ISB scientists collaborate with scientists from Center for Infectious Disease Research and Johns Hopkins University to identify new proteins on the surface of the malaria parasite Plasmodium.
The research aims to provide new targets for a malaria vaccine.
It is discovered that some Plasmodium surface proteins have sugar attachments that can cloak these proteins from the human immune system.

A Global Map To Fight Tuberculosis
The disease progression of tuberculosis is extremely complex and it’s poorly understood.
ISB and Seattle BioMed researchers have made an important step by developing a comprehensive map of gene regulation in tuberculosis.
A resulting open-access web portal offers any scientist the ability to mine the collected data.

A New Approach to Identifying How the Deadly Dengue Virus Multiplies
Dengue virus is the most prevalent mosquito-borne virus worldwide, infecting an estimated 400 million people per year and causing about 25,000 deaths.
It’s necessary to understand the molecular mechanisms of dengue replication in order to develop an effective treatment.
Researchers at ISB and Seattle BioMed developed a novel approach for identifying host proteins that associate with dengue replication machinery.

Uncovering the Genetic Adaptability of Tuberculosis
The Institute for Systems Biology and Seattle BioMed have collaborated to reconstruct the gene regulatory network of the human pathogen Mycobacterium tuberculosis.
Finely tuned gene regulation has allowed Mycobacterium tuberculosis to survive unnoticed in an apparently healthy host for decades; understanding those subtleties is critical for advancing treatment.
The identification of co-regulated sets of genes and their regulatory influences offers validated predictions that will help guide future research into Mycobacterium tuberculosis pathogenicity.
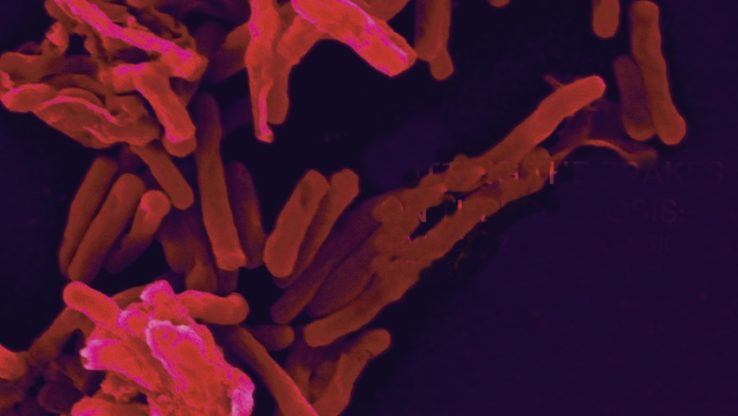
New Protein Modification Critical to Growth of Tuberculosis Pathogen Found
Institute for Systems Biology and Seattle BioMed researchers collaborated and discovered a new protein post-translational modification in the human pathogen Mycobacterium tuberculosis.
Post-translational modifications are essential mechanisms used by cells to diversify protein functions and ISB scientists identified the rare phosphorylated tyrosine post translational modification on Mycobacterium tuberculosis proteins using mass spectrometry.
Inhibiting phosphotyrosine modified amino acids in Mycobacterium tuberculosis severely limits the growth of this widespread deadly pathogen.
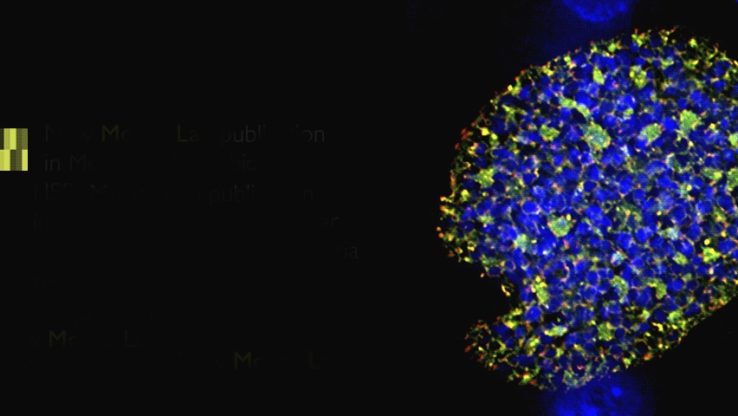
The Bug Stops Here: Arresting Malaria Parasites in the Liver Gives Immunity
ISB researchers collaborated with Seattle BioMed researchers to identify molecular building blocks required by malaria parasites to build cell membranes.
Deleting key genes necessary for building cell membranes created a parasite that does not make the host sick and can’t be passed on through the blood.
Treating mice with modified parasites gave them complete immunity against malaria.



