A Mixture of Markers from Two Distinct Cell Types Indicates Poor Prognosis in Breast Cancer
 isbscience.org/news/2015/06/15/two-distinct-cell-types-poor-prognosis-breast-cancer/
isbscience.org/news/2015/06/15/two-distinct-cell-types-poor-prognosis-breast-cancer/
3 Bullets:
- Identifying the most aggressive cells in cancer (cancer stem cells) is essential for designing effective therapy and predicting patient outcomes.
- Using single-cell analysis techniques, researchers at ISB and Luxembourg Centre for Systems Biomedicine have identified the molecular signatures of two types of malignant breast tissue cells.
- Researchers found an interesting twist: the two cell types “cooperate” to increase malignancy potential and they promote a third hybrid stem-cell type.
By Drs. Anne Grosse-Wilde and Sui Huang
Metastatic breast cancer remains an incurable disease and has stimulated the search for the most aggressive cell types in the tumors that drive metastasis. These cells have long been thought to possess stem-cell character, hence the idea of Cancer Stem Cells, or CSCs. In breast tissue, one can distinguish between two cell types: epithelial (E) and mesenchymal (M) cells that exert complementary functions. They both also exist in their cancerous equivalent. Thus a longstanding question has been: which one, the E-type or the M-type cancer cells represent the CSCs? The technique called “gene expression profiling” – the measurement of thousands datapoints indicating whether a cell’s genes are active or inactive (“molecular signatures”) – has become routine in the past decades and has been employed to identify the content of breast tumor tissue samples for activated “Egenes” or “M genes.” But results have been conflicting: some studies of breast cancer tissues suggest that predominance of E genes is associated with poor prognosis and enrichment for CSCs in the primary tumors, while others suggest that indeed, M genes (as traditionally believed) indicate poor outcome.
Journal: PLOS ONE
Title: Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival
Authors: Anne Grosse-Wilde, Aymeric Fouquier d’Hérouël, Ellie McIntosh, Gökhan Ertaylan, Alexander Skupin, Rolf E. Kuestner, Antonio del Sol, Kathie-Anne Walters, and Sui Huang
Link: PDF
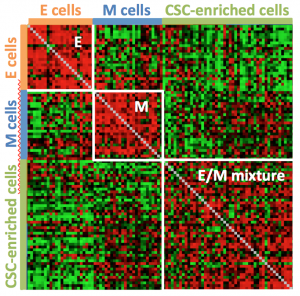
Correlation matrix of single cell gene expression data (every row/column represents a different cell) showing distinct patterns for E and M cell-types and a third distinct pattern for E/M cell types. Red pixel indicates high and green pixel low similarity between a pair of cells (correlation).
One problem with all these studies is that conventional expression profiling “grinds up” tumor samples in order to measure the molecular profiles, and thus, lack the resolution down to single cells. Rare, unknown cell types, including CSCs would escape such analysis. Now, the lead author of the study, Anne Grosse-Wilde, a cancer researcher and postdoc in the Ozinsky (formerly at ISB) and Huang labs, used the new technique of single-cell gene expression analysis to address the problem by analyzing E and M cells at single-cell resolution.
First, Dr. Grosse-Wilde generated several breast pure E and M cell-lines that enabled the researchers to identify gene expression signatures that specified E and M cells. Indeed, with this new technique of single-cell gene expression signatures she identified the E and M cell-types, but she also discovered a third distinct cell type: an intermediate between E and M, which was called “hybrid E/M cell-type”. Functional studies in cell culture revealed that E and M cell types were functionally opposite and complement each other with respect stem cell-associated properties. By contrast, the hybrid E/M cells displayed a combination of these stem cell-associated properties and thus exhibited full stem-cell functionality, such as survival in the floating culture, and notably, the ability to generate “mammospheres” – multi-cellular spherical structures observed in normal breast development. It is known that mammosphere-forming capacity is a hallmark of CSCs, and of aggressiveness when observed in malignant cells.
Intriguingly, mixing E and M cells in the same culture also generated mammospheres, and cells in such mixed cultures did so at dramatically increased efficiency. Thus, E and M type cancer cells, being complements, could cooperate to augment malignancy. Therefore, seeing an intermediate E//M molecular signature in the entire “grinded-up” tissue samples of patients could indicate two mechanisms of malignancy: first, the presence of E and of M cells which, as a mixture, can cooperate and second, the presence of actual individual hybrid E/M cells that carry CSC properties.
The researchers then confirmed their finding in clinical data: Using a large breast cancer database of more than 3,000 patients, breast tumors enriched for both the E and M signatures were indeed found in patients with poor outcomes, independent of the particular sub-type of breast tumors. Thus, the simplistic idea to find the culprit malignant cell in tumors that must be killed must be revised: these results suggest that we have to deal with the cooperation between the E and M cells, as well as to target the hybrid E/M cells. Since this result is independent of breast cancer type, this strategy holds the promise to improve survival in the majority of breast cancer patients.
This result also goes beyond breast cancer and teaches us that one cannot simply seek to pinpoint the cell type in a tumor that accounts for its aggressiveness. Like in many living systems, it takes two to tango.






