ISB’s Role in TCGA
 isbscience.org/news/2012/10/23/isbs-role-in-the-cancer-genome-atlas/
isbscience.org/news/2012/10/23/isbs-role-in-the-cancer-genome-atlas/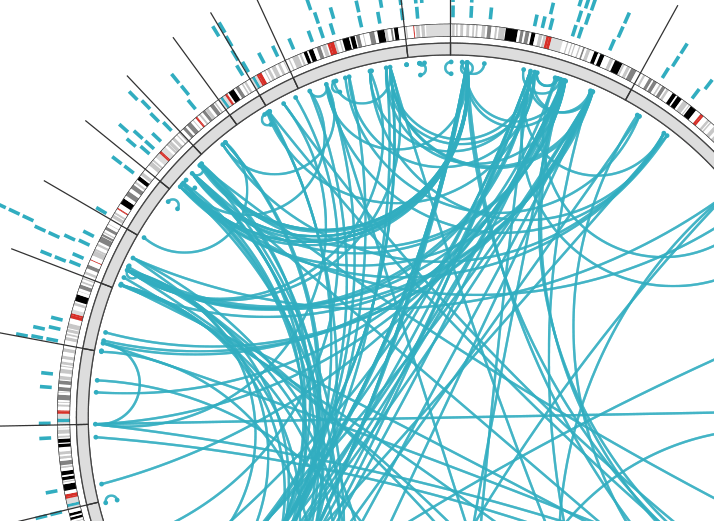
When you see a reference to “cancer research,” you know that it’s important. But do you really know what it means and how complex the research is? Many ISB scientists are entrenched in molecular cancer research. To better appreciate what they’re tackling, let’s talk about The Cancer Genome Atlas (TCGA) project.
In understanding cancers, researchers first have to know what errors in the DNA of tumor cells cause them to grow out of control. Being able to catalog the genomes – the entire DNA sequence – of each type of cancer cell helps researchers see where the errors occur and the context in which they take place. Once there are “maps” of these cancer genomes, scientists then can start to look for causal relationships. Eventually, this knowledge will lead to better prevention, detection and treatment for cancers.
Because there are more than 200 cancer types and subtypes, the process of collecting tissue samples and analyzing molecular as well as clinical data is a hairball of a task. But several hundred researchers from institutions across the country are collaborating on TCGA in order to characterize the complete genomes of more than 20 tumor types. Scientists in the Ilya Shmulevich group at ISB have contributed to TCGA research since 2009.
TCGA began as a three-year pilot project in 2006 with $50 million each from the National Cancer Institute and National Human Genome Research Institute. The pilot project proved that the work of collecting this type of tumor data was possible and that having an atlas of this information accessible to all would aid in new discoveries. It also demonstrated the effectiveness of engaging teams of inter-disciplinary scientists to advance a common cause.
ISB, in partnership with the MD Anderson Cancer Center, is one of the seven Genome Data Analysis Centers. GDACs are responsible for developing new technologies that help analyze and integrate more than a dozen disparate types of data, such as clinical diagnosis, treatment history, histological diagnosis, gene expression/RNA sequence, DNA sequence, DNA mutation, and miRNA expression.
The first paper published after the pilot project phase of TCGA – ”Comprehensive molecular characterization of human colon and rectal cancer” – appeared in the July 19 issue of Nature and also was featured in a cover story in the New York Times. ISB senior research scientist Vesteinn Thorsson, a member of the Shmulevich group, explains the team’s work:
“Collaborating with Timothy A. Chan, MD, PhD, of the Memorial Sloan-Kettering Cancer Center, we identified a set of six clinical measurements that are indicative of aggressive colorectal cancer. This includes whether the tumor has found its way into the bloodstream or into neighboring lymph nodes, whether the tumor has metastasized, and what stage the tumor has reached. We scored each of the thousands of molecular features available to us in the TCGA data set to indicate the degree of association with tumor aggressiveness. The score results from combining results from individual statistical association tests (p-values) between molecular variable and the six clinical variables using a weighted form of Fisher’s method. This yielded ranked lists of genes whose expression may be indicative of aggressive or less aggressive tumors. It also yielded corresponding lists for other kinds of colorectal tumor data generated by TCGA: DNA mutations, DNA copy number variation, methylation of gene promoters, and the expression of microRNAs.”
One of ISB’s unique contributions to TCGA is a Web-based tool called Regulome Explorer, which facilitates the integrative exploration of associations in clinical and molecular data from TCGA. Regulome Explorer comprises four applications, including the CRC Aggressiveness Explorer featured in the aforementioned colorectal cancer paper.
The importance of ISB’s Regulome Explorer can’t be stressed enough. This suite of tools has allowed TCGA researchers to easily visualize massive amounts of heterogeneous (different) data in a format that has revealed relationships that otherwise wouldn’t have been discovered. If you’ve ever had to compare and collate information from multiple tabs of a spreadsheet file, you have just an inkling of what TCGA scientists face. Processing a single dataset takes the equivalent of 5,000 dual-core computers about an hour to complete. That’s a lot of computation.
We are also proud to share that Google featured a version of one of our Regulome Explorer applications during a keynote presentation at the Google I/O developers conference in June.
In future issues of Molecular Me, we will share more reports on ISB’s work with TCGA, including the recent paper related to breast cancer findings that was also published in Nature.
Watch ISB senior research scientist Vesteinn Thorsson explain ISB’s TCGA work.





