ISB is pioneering global health innovations for a safer future.
We are exploring new approaches that focus on cutting-edge treatments and preventive measures to fight infectious diseases and tackle today’s most pressing global health challenges.
Our infectious disease research addresses a wide range of global health initiatives.

Dr. Anna Kuchina, center, and members of her lab, working at ISB. Image credit: Scott Eklund/Red Box Pictures.
We are exploring new approaches that focus on cutting-edge treatments and preventive measures to fight infectious diseases and tackle today’s most pressing global health challenges.

Dr. Nitin Baliga in the lab at ISB. Image credit: Scott Eklund/Red Box Pictures.
“Tuberculosis, malaria, and other infectious diseases are global scourges. Systems biology enables us to develop better diagnostics and more effective vaccines and treatments for these deadly diseases, which disproportionately affect the world’s poorest populations.”
Dr. Nitin Baliga, ISB SVP, Director and Professor
Fighting antimicrobial resistance and advancing alternative therapies
Accelerating breakthroughs in detection, treatment, and resistance
Advancing vaccines to block transmission and improve global health
Developing new therapies and vaccines to combat a wide range of infections
Uncovering treatments and improving diagnostics for better outcomes
Decoding the role the immune system plays in symptoms and treatment
We are taking on the growing threat of drug-resistant infections by exploring new therapies and alternative solutions. By studying how bacteria and viruses evade current treatments, we hope to develop innovative approaches that will provide more effective options for combating antimicrobial resistance.
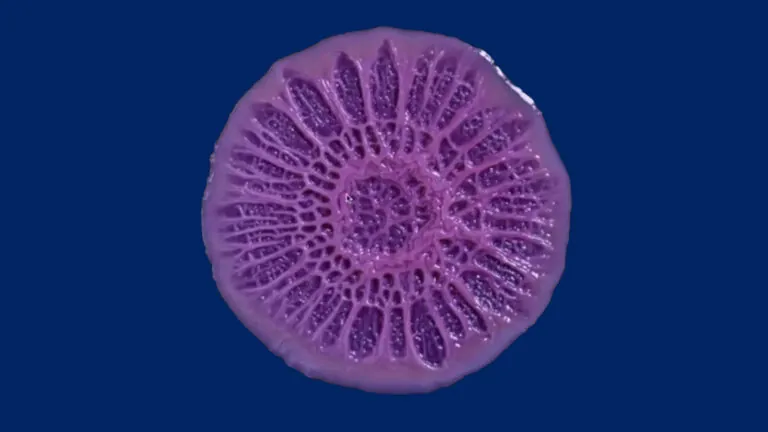
Still from a time-lapse video of growing Pseudomonas aeruginosa biofilm. Image credit: Scott Chimileski.
How bacteria – including infectious ones – coalesce into assemblages called biofilms that help them resist antibiotics is not well understood. Assistant Professor Anna Kuchina invented a technology (microSPLiT) that allows researchers to individually study hundreds of thousands of bacteria in a single experiment. The recipient of a promising investigators NIH grant (MIRA), her Lab focuses on closing the gap in our understanding of bacterial biofilms.
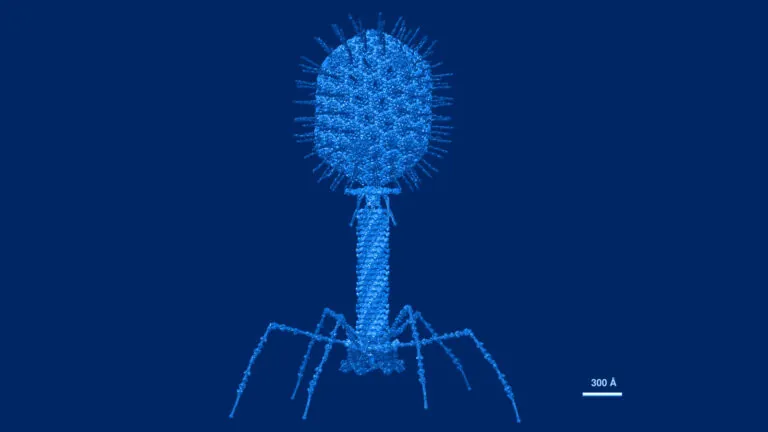
Atomic structural model of bacteriophage T4 in UCSF Chimera software using pdbs of the individual proteins. Image credit: Victor Padilla-Sanchez, PhD. CC BY-SA 4.0. Image has been recolored by ISB.
Antibiotics – the most common treatment for bacterial infections – deplete many species in the gut microbiome and can lead to resistance. Phage therapy, an alternative to antibiotics, relies on viruses called bacteriophages that each target only one type of bacteria. To potentially improve phage therapies, the Kuchina Lab is investigating how gut microbiota species defend against bacteriophage attacks.
Stock illustration of antimicrobial resistance. Credit: AdobeStock.
All organisms – from humans to microorganisms – utilize adaptive prediction to anticipate and prepare to deal with future environmental challenges. This gives infectious disease microorganisms – including bacteria and viruses – an opportunity to develop AMR to drugs, a catastrophic problem for global health. The Baliga Lab, with support from the NIH, is developing ways to disrupt adaptive prediction and resulting AMR.
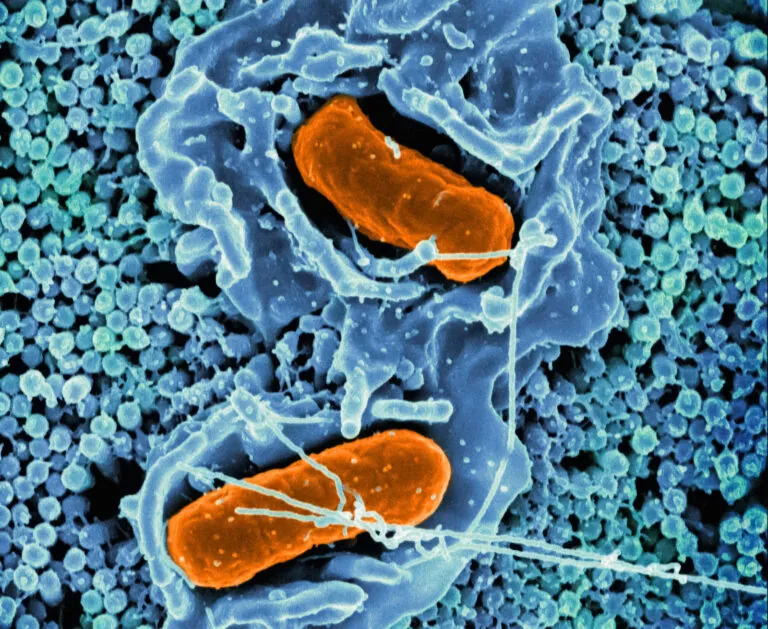
Scanning electron micrograph of Salmonella Typhimurium invading a human epithelial cell. Image credit: NIAID, recolored by ISB.
The Centers for Disease Control estimates there are 1.35 million U.S. Salmonella infections annually, with infants, older adults, and immunocompromised individuals most at risk for severe infections. The Subramanian Lab, with NIH funding, has discovered that salmonella “cross-talks” with our immune system to conceal itself and resist treatment. These insights can inform the development of therapies to combat this emerging drug resistance threat.

Dr. Jie Luo working at ISB. Photo credit: ISB.
Viral global pandemics affect all of us. Drugs that can be cheaply and rapidly deployed in the early stages of outbreaks – even before vaccines are available – are critical. The Ranish Lab, with Washington Research Foundation support, created a new broad-spectrum anti-viral compound that holds the promise of being quickly deployable, effective, and inexpensive.
Our scientists are leading the way in tuberculosis research to enhance detection, treatment, and prevention. Through advanced approaches, we aim to speed up drug discovery, develop faster and more effective treatment regimens, and create affordable diagnostic tools. These efforts are crucial steps in the quest to combat drug-resistant TB and reduce the global burden of the world’s deadliest infectious disease.
Tuberculosis bacteria. Image credit: CDC.
The pathogen Mycobacterium tuberculosis (Mtb) that causes TB is adept at gaining resistance to treatment, making it difficult to eliminate. The World Health Organization has declared multi-drug resistant TB (MDR-TB) a global health crisis. The Baliga Lab, with NIH funding, is accelerating the development of multidrug regimens to achieve fast and complete clearance of Mtb and reduce MDR-TB.

Image of prescription TB drugs. Photo credit: Stockvault, recolored by ISB.
The global TB epidemic is worsening as new TB strains emerge that are resistant to current drug treatments. Standard TB treatments are 3-9 months, which for many patients is impractical. The Baliga Lab, with support from the Gates Foundation, is discovering new drug targets and uncovering novel mechanisms of action for next-generation TB agents that can shorten treatment and prevent TB drug resistance.

Patient samples. Photo credit: Victoria Uhl /ISB.
The World Health Organization estimates that up to a third of the world’s population may be latently infected with TB bacteria, with 5-10 percent of these people developing active, transmittable TB. There is a pressing need for low-cost tests to screen for active TB. The Baliga Lab has developed a new blood-based method to diagnose active TB pre-symptomatically at a very early stage and is assessing ways to develop a low-cost screening tool.

Dr. Nitin Baliga in the lab at ISB. Photo credit: Scott Eklund / Red Box Pictures.
Following exposure to TB, the constantly evolving combination of genetic differences in humans and in the Mtb bacteria itself results in a wide array of patient outcomes. To improve clinical decision-making, the Baliga Lab is working with the University of Washington to understand these complex genetic interactions, how they affect TB patients, and the best treatment options.

By using a computer model to understand the adaptions of Mycobacterium tuberculosis (Mtb), the pathogen that causes tuberculosis, researchers at ISB have identified a network within Mtb that allows it to tolerate and resist drug therapies. This work is published in Cell Reports.

Tuberculosis (TB) is the world’s second leading infectious disease killer after COVID-19. Drug resistance to TB is a public health crisis. ISB researchers have developed algorithms to predict the efficacy of drugs in treating Mycobacterium tuberculosis (MTB), the causative agent for TB. These research findings were published in the journal Cell Reports Methods.

ISB researchers Dr. Nitin Baliga, Dr. Eliza Peterson and Dr. Vivek Srinivas have developed a new cell sorting technology, called PerSort, that isolates and characterizes dormant persisters that exist in cultures of Mycobacterium tuberculosis, the pathogen that causes tuberculosis.
ISB researchers are focused on improving the effectiveness of malaria vaccines and developing ways to prevent the spread of the disease. Our goal is to contribute to better health outcomes worldwide by supporting global efforts to reduce malaria’s impact and protect vulnerable communities.
Close up of lab work at ISB. Photo credit: Scott Eklund / Red Box Pictures.
Malaria vaccines offer limited protection. These vaccines target proteins on a parasite that transmits malaria to humans by mosquito salvia. Kristian Swearingen in the Moritz Lab, with Seattle Children’s Research Institute, is developing a deeper understanding of these parasite proteins to help meet the World Health Organization’s goal of having a vaccine with a protective efficacy of at least 75 percent.

View of a mosquito under a microscope. Image credit: Dr. Kristian Swearingen/ISB.
Of the two vaccines against malaria, both have less than 50 percent efficacy against severe illness and don’t prevent transmission of the parasite that causes malaria as it cycles from mosquitos to humans. Kristian Swearingen in the Moritz Lab, with Seattle Children’s Research Institute, is accelerating vaccine development by identifying transmission-blocking parasite target proteins.
ISB researchers are leading efforts to develop innovative therapies and vaccines for combating influenza and other serious lung infections. We hope to create more effective treatments, reduce the impact of infections, and protect the health of individuals worldwide, especially those most vulnerable to severe illness.

Electron micrograph of influenza virus. Image credit: Wikimedia Commons, recolored by ISB.
Secondary bacterial infections are a major cause of severe illness and death from influenza. People over 65, children under 5, and people with certain chronic conditions are especially at risk of serious illness. Kathie Walters of the Hood Lab works with researchers from the National Institute of Allergy and Infectious Disease to identify targets for potential new therapies for severe influenza-bacteria co-infections.
Syringe with needle and vaccine. Photo credit: Allison Saeng, Unsplash, recolored by ISB.
The Centers for Disease Control recommends an annual flu shot, the composition of which is updated each flu season, for everyone over 6 months. Kathie Walters of the Hood Lab is a long-time collaborator with the National Institute of Allergy and Infectious Disease and is working to ensure that vaccines against influenza viruses are as effective as possible.
A 3D-generated image of Streptococcus pneumoniae. Image credit: CDC, recolored by ISB.
Pneumonia from an infection by the bacteria Francisella tularensis (tularemia pneumonia) from a tick or deer fly bite, if left untreated, has a mortality rate of up to 60 percent. With funding from the NIH, the Hood Lab’s Kathie Walters and Harborview Medical Center are studying how these bacteria evade lung defenses to identify targets for Francisella vaccine development, which may help understand other lung infections.
We are at the forefront of the global effort to understand and treat COVID-19 and Long COVID. From improving diagnostic tests to determining which patients will be more likely to experience severe Long COVID, our scientists are dedicated to developing more effective treatments and prevention strategies. These efforts are vital to enhancing recovery and outcomes for individuals worldwide.

RECOVER Consortium branding image. Image credit: RECOVER/ISB.
In 2023, the CDC estimates that 7 percent of U.S. adults had Long COVID, which is debilitating and difficult to treat. Jim Heath is leading the Pacific Northwest Consortium of the NIH-funded RECOVER nationwide Long COVID study. The Consortium includes Swedish, Providence Spokane, UW, and Cedars-Sinai and is working to unravel the confounding heterogeneity of Long COVID and to unearth potential treatments.

Dr. Jenn Hadlock with her lab at ISB. Photo credit: Scott Eklund / Red Box Pictures.
At least 10 percent of adults with COVID-19 will experience Long COVID with a wide range of health problems extending at least three months after initial infection. The Hadlock Lab, with NIH support, is exploring how our immune system’s adaptive response to COVID-19 infection differs in patients with Long COVID to pinpoint potential prevention and treatment strategies.

Drs. Inyoul Lee and Kelsey Scherler at ISB. Photo credit: Steve Utaski / Remedy Pictures.
COVID-19 remains a serious health issue. In 2023, it was the 10th leading cause of U.S. deaths. Kai Wang and Inyoul Lee of the Hood Lab, with NIH support, are assessing the validity of a new diagnostic test developed by The Ohio State University. The test uses saliva or blood rather than invasive nasal swabs and can provide more detailed diagnostic information.

Cover image from Cell issue featuring ISB Long COVID research. Image credit: ISB.
Researchers have identified several factors that can be measured at the initial point of COVID-19 diagnosis that anticipate if a patient is likely to develop long COVID. They also found that mild cases of COVID-19, not just severe cases, are associated with long COVID. Their findings were published by the journal Cell.

Image credit: Joshua Bright / The New York Times.
In an article published by Pam Belluck for the New York Times titled, “New Research Hints at 4 Factors That May Increase Chances of Long Covid,” findings of an ISB-led study published in the journal Cell are covered in depth, including quotes from ISB president Dr. Jim Heath.
At ISB, we are dedicated to unraveling how the immune system responds to this tick-borne illness with the goal of improving diagnosis and treatment. By studying the body’s reaction to Lyme disease, our researchers aim to develop more effective solutions for preventing long-term symptoms and improving quality of life for those who contract it.

Hand holding a glass tube containing a tick. Photo credit: Marino Linic, Unsplash, recolored by ISB.
Lyme disease, with 500,000 annual U.S. cases, cannot be quickly diagnosed and can become disabling. The Subramanian Lab, with DoD funding, is tracking the immune response to Lyme disease in patients over time to define molecular and cellular transitions in acute disease and understand why some individuals progress to long-term disease. These insights are critical for improving diagnosis and treatment.

“Our lab is focused on understanding why some individuals can clear Lyme disease with antibiotic treatment, while others progress to more serious, chronic conditions such as neurological, cardiac, or joint complications. We believe that insights into the immune system’s response will pave the way for improved diagnosis and more effective treatment options.”
Dr. Naeha Subramanian, ISB Associate Professor