Sept. 8, 2015
3 Bullets:
- General transcription factor TFIIH plays central roles in gene transcription and DNA repair
- ISB researchers and collaborators map the architecture of the TFIIH complex using powerful crosslinking-mass spectrometry (CXMS) technology and integrative modeling
- Structural maps provide critical insights into how mutations in TFIIH subunits lead to disease phenotypes
By Jie Luo and Mark Gillespie
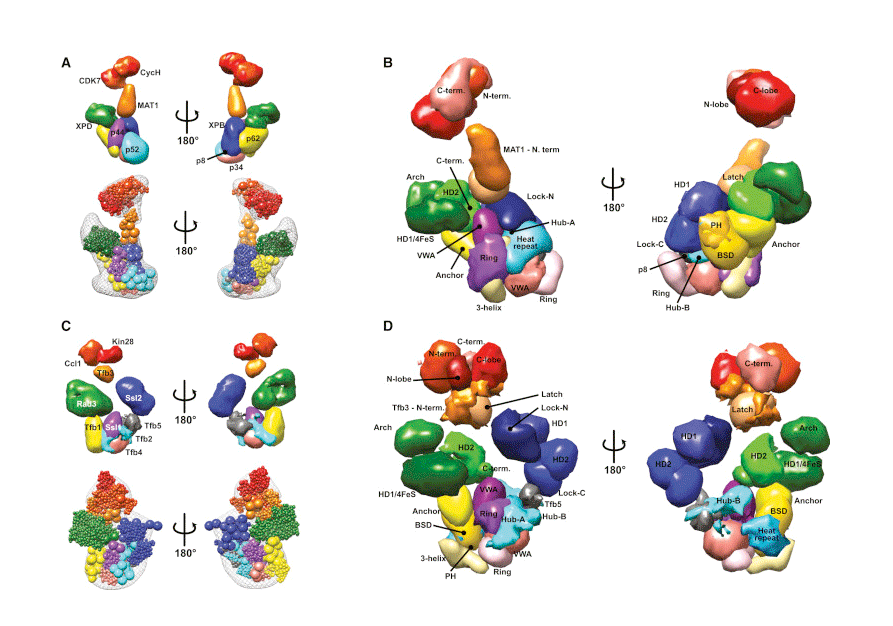
The expression, or transcription, of genes controls the identity and function of a cell. DNA damage caused by UV light or other carcinogens must be repaired to maintain genome integrity. The general transcription factor TFIIH plays central roles in both processes and is also important to couple gene transcription with DNA repair. Researchers at the Institute for Systems Biology, in collaboration with the University of California, San Francisco, the University of Colorado Boulder, and the Fred Hutchinson Cancer Research Center, have mapped the architecture of the multi-subunit TFIIH complex. This research, published online in Molecular Cell on Sept. 4, 2015, represents a breakthrough in understanding the structural basis for transcription and DNA repair, and provides critical insights into how disruption of the TFIIH complex can lead to cancer and other diseases.
Journal: Molecular Cell
Title: Architecture of the human and yeast general transcription and repair factor TFIIH
Authors: Jie Luo, Peter Cimermancic, Shruthi Viswanath, Christopher C. Ebmeier, Bong Kim, Vishnu Raman, Charles H. Greenberg, Riccardo Pellarin, Andrej Sali, Dylan Taatjes, Steven Hahn & Jeff Ranish
Link: read paper
TFIIH is a large multi-subunit protein complex, with flexible domains and numerous conformations, which hindered previous attempts to delineate its structure. ISB researchers Jie Luo and Jeff Ranish developed a powerful crosslinking-mass spectrometry (CXMS) approach and applied their technology to TFIIH. With CXMS, protein-protein interactions are efficiently captured using chemical crosslinkers and identified using state-of-the-art mass spectrometry. By performing their experiments on both human and yeast TFIIH, Luo and Ranish identified evolutionarily conserved crosslinks, which they used to deduce both the spatial organization and conserved protein domain interactions within the complex. This comparative CXMS strategy provided high quality spatial information to collaborators at UCSF, who subsequently used their integrative modeling platform to assemble a detailed structural model of the TFIIH complexes. From this, four new conserved “topological regions” and more than 35 conserved protein domain interactions within TFIIH, were identified, thereby illuminating a network of interactions involved in TFIIH assembly and regulation of its activities.
Mutations in TFIIH subunits are associated with many forms of cancer and autosomal recessive disorders, such as Xeroderma Pigmentosum (XP), Tricothiodystrophy (TTD), and the combined symptoms of XP and Cockayne syndrome (XP/CS). Interestingly, most of the mutations found in patients do not directly affect the enzymatic activities of TFIIH, but rather the interactions between the enzymatic subunits and their regulatory partners. By determining the architecture of large protein complexes, researchers can identify mechanisms explaining how mutations lead to disease phenotypes, and identify potential targets for therapeutic intervention.


 isbscience.org/research/researchers-determine-architecture-of-a-macromolecular-complex-regulating-gene-expression-and-dna-repair/
isbscience.org/research/researchers-determine-architecture-of-a-macromolecular-complex-regulating-gene-expression-and-dna-repair/