Jan. 12, 2014
3 Bullets:
- Price Lab researchers worked with collaborators at the University of Illinois to create an easy-to-use software toolkit for comparing microbial genomes.
- Tools can be used to find orthologs, correct missing/inaccurate gene annotations, analyze gene gain and loss patterns, and build draft metabolic networks from reference networks.
- Software offers predictive and analysis capabilities in a flexible way, making it easy to build customized analysis pipelines.
By Matt Richards
The recent boom in DNA sequencing has resulted in a large quantity of data and has enabled comparative studies of the genetic content of many related species. Researchers in the Price Lab at the Institute for Systems Biology and their collaborators at the University of Illinois have developed new software — Integrated Toolkit for Exploration of Microbial Pan-Genomes, or ITEP — designed to expedite comparative studies by combining erstwhile separate tools in one workspace. The study was published on Jan. 4 in the journal BMC Genomics.
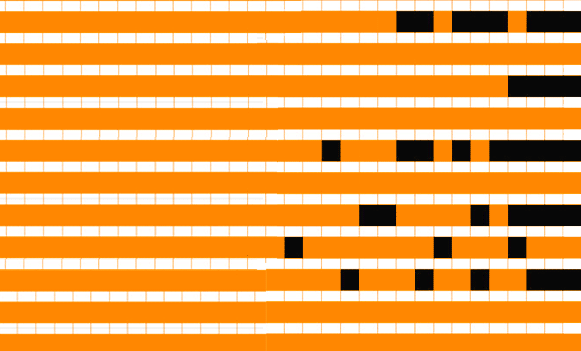
Analysis of related gene families provide a window into how genes are passed down over the course of evolution and give us insight into common protein functions between species. ITEP includes tools to align sequences, identify orthologs, check for missing gene calls, visualize cluster integrity, and analyze phylogenetic information used to examine evolutionary relationships. This allows researchers to rapidly identify patterns in genetic variation across related species and cross-reference these to the organism phylogeny or known physiological differences to gain insight into the genetic basis for this variation.
As with all ISB software, ITEP is open-access and can be downloaded at http://price.isbscience.net/itep. To read the full journal article, visit http://www.biomedcentral.com/1471-2164/15/8.


 isbscience.org/research/powerful-new-program-integrates-multiple-tools-into-a-flexible-interface/
isbscience.org/research/powerful-new-program-integrates-multiple-tools-into-a-flexible-interface/