Seeing the Pathogen’s Perspective: ISB Researchers Develop Method to Profile Pathogen Gene Expression from Infected Host Cells
 isbscience.org/news/2019/03/04/seeing-the-pathogens-perspective-isb-researchers-develop-method-to-profile-pathogen-gene-expression-from-infected-host-cells/
isbscience.org/news/2019/03/04/seeing-the-pathogens-perspective-isb-researchers-develop-method-to-profile-pathogen-gene-expression-from-infected-host-cells/- Researchers at Institute for Systems Biology (ISB) have developed a method, Path-seq, that simultaneously profiles host and pathogen gene expression and is sensitive enough to explore the pathogen transcriptome in vivo.
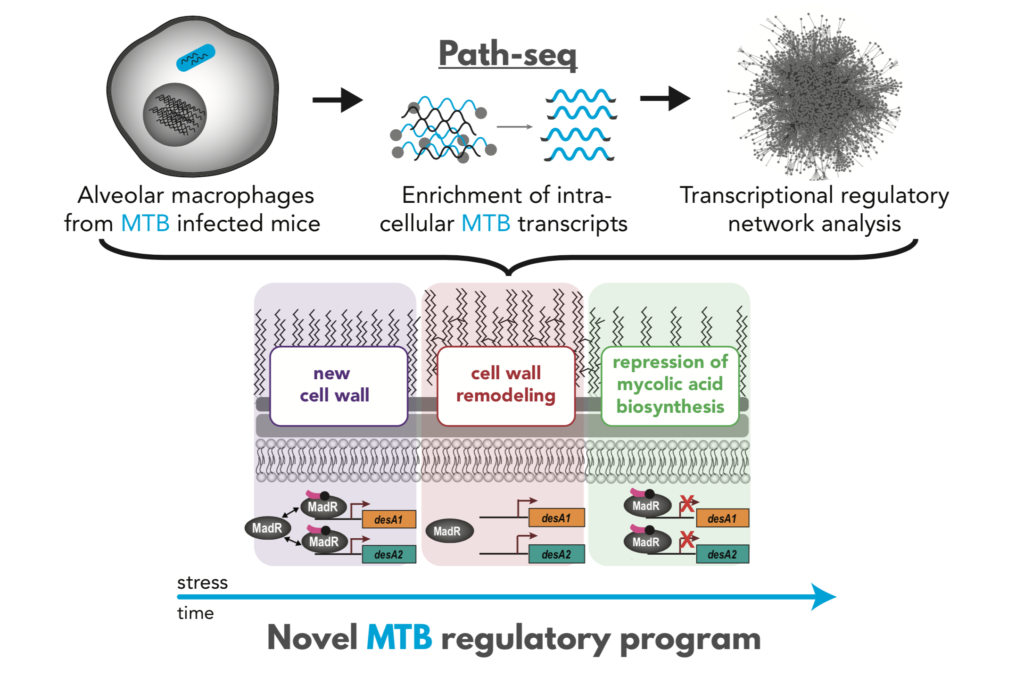
- Using both Path-seq data from the human pathogen Mycobacterium tuberculosis and systems approaches, the study revealed infection-specific regulatory networks, including a network that plays a role in the composition and levels of cell wall mycolates.
- Path-seq represents a significant advance in understanding the authentic intracellular context experienced by pathogenic bacteria, thus informing novel strategies to fight tuberculosis and other infectious disease.
The rise in multi-drug resistant (MDR) and extremely drug resistant (XDR) strains of Mycobacterium tuberculosis (MTB) has become a major cause of global health concern for treating tuberculosis. The number of worldwide deaths caused by tuberculosis has surpassed HIV/AIDS, stressing the sense of urgency for finding effective new drugs to fight MTB.
In a landmark study published today in Molecular Systems Biology, researchers at ISB, Seattle Children’s Research Institute, and University of Birmingham (UK) reported a novel method, Path-seq, to profile expression of all MTB genes within alveolar macrophages of infected mice. This study presents the most comprehensive transcriptome profiling of MTB from in vivo infection and a major technical advancement for studying any host-pathogen interaction.
“The key to MTB pathogenesis is largely its ability to survive within macrophages. Despite this importance, we have limited understanding of the molecular changes that occur in MTB during infection of these host immune cells, especially in vivo,” emphasized Dr. Eliza Peterson, senior research scientist in ISB’s Baliga Lab and first author on the paper. Technical challenges, primarily a striking excess of host over bacteria RNA, have limited the transcriptional profiling of pathogens during infection of animal models.
Previously, the Baliga lab published a whole cell gene network model that could predict how MTB senses and responds to changes in its environment. In this study, they used the network model and Path-seq data to understand how MTB responds to the intracellular environment within alveolar macrophages. They used this network-enabled knowledge to identify infection-specific regulatory networks and demonstrated that a network controlled by MadR (named for mycolic acid desaturase regulator) dynamically alters cell wall mycolates during infection.
“The ability afforded by Path-seq to ‘see’ the world from the pathogen’s perspective will enable strategies to target intracellular tuberculosis bacteria,”said Dr. Nitin Baliga, the senior author on the paper.
Indeed, the group went on to show that perturbation of MadR leads to MTB killing, confirming MadR as an attractive drug target. The success of Path-seq for observing and interpreting gene expression changes relevant to infection, demonstrates its potential to accelerate efforts towards shortening TB drug regimen.