Our Genomes, Our Selves: The hunt for a disease gene
 isbscience.org/news/2012/03/20/our-genomes-our-selves-the-hunt-for-a-disease-gene/
isbscience.org/news/2012/03/20/our-genomes-our-selves-the-hunt-for-a-disease-gene/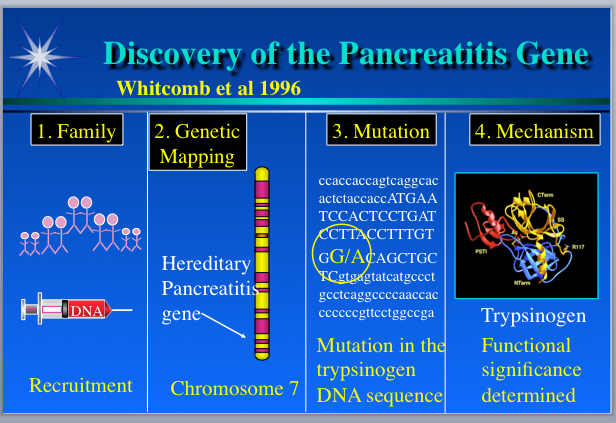
By Lee Rowen
Every once in a while, different components of a research problem come together at just the right time. One lives for, and savors, these occasions. One of my favorite examples is the discovery of the genetic variant that causes hereditary pancreatitis.
Hereditary pancreatitis (HP) causes painful attacks of the gut, often beginning in childhood. Over time, the pain and damage can become chronic and there is an increased risk for developing pancreatic cancer. It’s a rare disease that runs in families similar to the sorts of conditions we currently are investigating in the family genomics group at ISB (Hood Lab).
Although the methods used in 1996 to find the gene for HP have been replaced by the whole genome sequencing approach we use now at ISB, the underlying logic for how to find a disease-causing variant is the same.
In the mid-’90s, only a couple percent of the human genome had been sequenced, so people used laborious mapping techniques to locate the neighborhood of the genome where a detrimental genetic variation seemed most likely to reside. So-called sequence-tag-sites, which were small snippets of sequence with an approximately known chromosomal location, were used as markers. Using sets of these markers spread across the whole genome, disease-gene hunters would probe DNA samples from affected and unaffected individuals, with the aim of finding a marker in which a variation in the tag sequence would be identical for all of the folks with the symptoms, and dissimilar among people without symptoms. Because of the way inheritance of genomic DNA works, one can usually infer that the identified marker and the disease gene map are close to each other on the chromosome, and so that is where it makes sense to look for the problematic gene. To apply this approach to any given disease, researchers need a good set of DNA samples, which are often hard to acquire.
'Slone Disease'
But, in the case of HP, Bobby Slone, whose child showed severe symptoms, emerged as a hero. Slone was aware that this rare disease was not so rare in his extended family. He spoke to relatives and compiled a pedigree spanning nine generations, with information about who had the same set of peculiar symptoms. He brought this pedigree into his child’s doctor’s office in hopes it would prove helpful. Fortuitously, at around the same time, a group at the University of Pittsburgh had contacted the same doctor, asking him to be on the alert for patients with presumed HP because they were trying to identify the causative gene. A collaboration was born.
In 1996, Slone organized a family reunion picnic and asked those who were willing to donate blood. He collected about 90 samples. The Pittsburgh team then quickly used the sequence tag marker set to discover that the causative gene resides near the end of the long arm of chromosome 7.
Here’s where I enter the story. We had sequenced a chunk of that region on chromosome 7, because it contains immune system genes we were studying. Back in the early ’90s, a goal in Lee Hood’s lab was to figure out the best strategies for sequencing long contiguous stretches of human genomic DNA. We’d picked a difficult area, the immune T cell receptor (TCR) repertoire, where it was believed that many portions of the sequence are duplicated at high sequence similarity. Hence, it would be a huge challenge to sort out the puzzle correctly with methods that we had to develop. But we were up to it.
In 1994, after three years of intense effort, we deposited the first fully annotated human DNA sequence longer than 500,000 bases into GenBank, the public data repository, whereupon it immediately broke everybody’s software. Much to our surprise, within the T cell genes, we found five duplicated stretches of sequence containing a gene family called trypsinogen. Wow, genes inside genes. We had no idea.
A Gene is Found
Back at Pittsburgh, now that they knew where in the genome to look, the researchers employed what is called a “candidate gene approach” to discover the cause of HP. They asked: What genes known to map on the end of the long arm of chromosome 7 might have something to do with the pancreas or the gastrointestinal tract? They identified a handful of candidates, one of which was trypsinogen. Trypsinogen, produced in the pancreas, is the precursor of trypsin, an enzyme able to digest protein, including itself. They decided to search GenBank to see if the trypsinogen gene had been sequenced and whoa, a humongous sequence that contained several similar copies of trypsinogen appeared. It was our sequence.
The Pittsburgh team emailed me for advice. I helped them design ways to discriminate among the copies so they could test each trypsinogen gene separately using the blood samples they had from the Slone clan. It turned out that an ancestor had a sequence variation causing an amino acid change in one of the trypsins produced by trypsinogens. This mutant trypsin protein cannot be processed properly so it digests the wrong things, wreaking havoc in the digestive system. The Pittsburgh team had their gene and I got invited to present at the First International Symposium on Hereditary Pancreatitis in 1997.
Unfortunately, there is no cure for HP yet. But there is now a genetic test, which raises several ethical and social issues I will talk about in a subsequent post.
The HP story illustrates key elements of discovery that carry over into our work in the family genomics group at ISB. First is the generosity of families willing to donate their DNA and medical histories to a research effort aimed at finding a genetic cause for a disease. Second is having an approach in hand that can discern patterns of genetic difference between affected and unaffected individuals. And third is a collaborative environment in which data are freely shared among researchers, clinicians and, in some situations, the patients and families.
When these three elements are present, significant progress can occur.
Related links:
About Lee Rowen: Lee is a Senior Research Scientist in Dr. Leroy Hood’s lab. Her pedigree includes PhDs in biochemistry from Stanford University and philosophy from Vanderbilt University. She participated in the Human Genome Project between 1990 and 2003, sequencing immune receptor loci and large portions of chromosomes 14 and 15. She achieved her fifteen minutes of fame by winning the “GeneSweep” bet for predicting the number of genes in the human genome. She is now entering the world of personal genomics.






