A New Tool to Predict Tumor Drug Resistance
The Wei Lab’s MetaboCore uses a simple needle biopsy and analyzes tumor metabolism to quickly find cancer drug resistance and detect immune responses predictive of immunotherapy success, speeding up the process of finding an effective treatment. Early clinical studies in ovarian, colorectal, and bile duct cancers show MetaboCore to be a fast and reliable diagnostic tool.
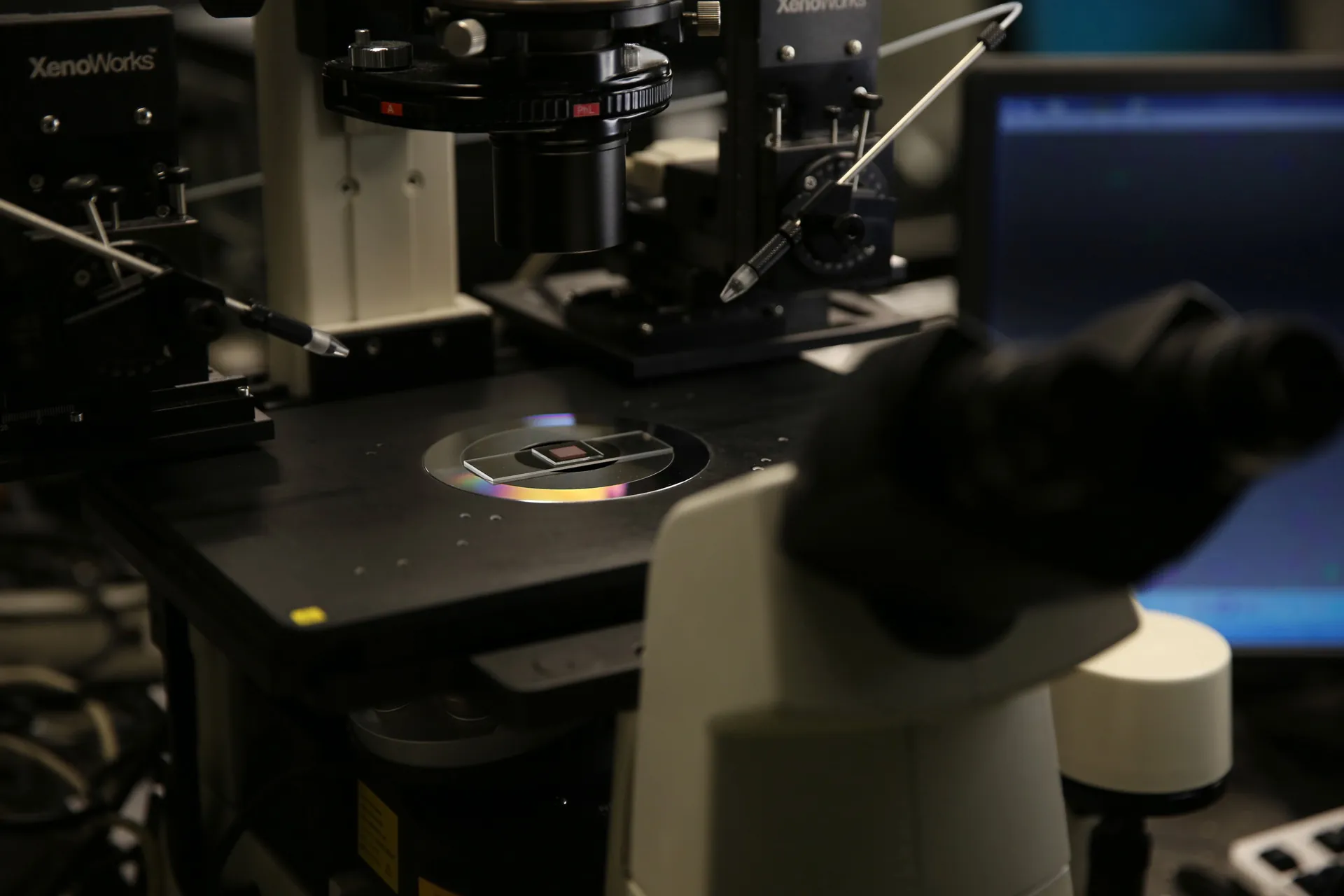
Photo of the single-cell on-chip metabolic cytometry device in use in the ISB optic lab. Photo credit: ISB.
Recently, physicians and scientists have come to appreciate that every cancer patient’s disease is unique. Although precision oncology holds the potential to match tailored treatments to individual patients, knowledge of how to pair the best treatment to each patient is often lacking. An ISB team has developed a new method, MetaboCore, that could quickly and affordably evaluate existing drugs in patient tumor biopsies, giving real-time information to physicians to inform precision cancer treatment. MetaboCore marries two unique methods developed by researchers at ISB and the University of Washington to detect changes in cancer cells’ metabolism as an early read-out of their sensitivity to cancer drugs.
- Funded by the National Cancer Institute, the Washington Research Foundation, Andy Hill CARE, and the Kuni Foundation.
- Led by Wei Wei, PhD
- Key collaborators:
- Raymond Yeung, MD, University of Washington
- Charles W. Drescher, MD, Swedish Cancer Institute
MetaboCore: Real-time drug sensitivity testing for cancer
MetaboCore is a treatment-evaluation method known as a drug-sensitivity assay. Previously developed drug-sensitivity assays have failed to show significant benefit to patients and to date, none have been widely adopted for clinical use. MetaboCore aims to transform past assays by integrating cancer cells’ natural environment, known as the tumor microenvironment, into the drug sensitivity testing. The tumor microenvironment includes many different kinds of non-cancerous cells and can play a large role in cancer’s survival or sensitivity to treatments.
MetaboCore works by taking microscopically thin slices of a tumor needle biopsy and keeping those slices alive in the lab through a method known as organotypic tumor culture, developed by Dr. Raymond Yeung at the University of Washington. The slices are then treated with cancer drugs, allowing testing of up to 10 different drugs from three small needle biopsies, a more-than-sufficient number to guide oncologists’ treatment decisions. After treatment, the slices are then broken down to individual cells and run through an assay developed by ISB’s Dr. Wei Wei, known as on-chip metabolic cytometry, that tests the cells’ metabolic response to the treatment.
These tiny chips are loaded with fluorescent markers that indicate different aspects of the metabolism of the cancer cells and of the healthy cells in their microenvironment within the tumor slice. Wei and his colleagues look for indications that the cells are reducing their use of glucose for energy, a process known as glycolysis. Lowered glycolysis is an early indicator that a given drug is working to kill cancer cells. MetaboCore can also detect other kinds of metabolism shifts that reveal important information about the health of the individual cells in the assay. For example, it can identify immune cell activation, characterized by increased glycolysis and oxidative stress, which is a critical signature associated with positive immunotherapy responses.
Because MetaboCore tests drugs directly on biopsy samples, as compared to other assays that grow cancer cells in the lab or recreate patients’ tumors in mice before testing them, the assay can return results within a week, quickly enough to guide patient treatment decisions in real-time. And because the assay uses cutting-edge single-cell approaches, it can be performed on less than 10,000 cells from a biopsy, a much lower number than is needed for other forms of drug-sensitivity assays.
Wei and his colleagues have tested an earlier version of their metabolic read-out method on samples taken from lung cancer patients and found that their tests predicted patients’ response to treatment with 95% accuracy, more accurately than the standard-of-care imaging methods. The team is now testing MetaboCore on samples from patients with liver cancer, colorectal cancer, ovarian cancer, and cholangiocarcinoma, and will expand to endometrial cancer and melanoma. Early results from colorectal and cholangiocarcinoma cancer patients have also shown high levels of accuracy in predicting patient response to treatments. The team expects that the method can be used for any solid tumor type. Currently, the method can assess chemotherapy drugs and targeted biologic drugs; the team is working to expand the method so that it can be used to test immunotherapies as well.
Citations
- Li, Z., et al. Liquid biopsy-based single-cell metabolic phenotyping of lung cancer patients for informative diagnostics. Nat Commun. 2019. doi: 10.1038/s41467-019-11808-3.
- Yang, et al. Hexokinase 2 discerns a novel circulating tumor cell population associated with poor prognosis in lung cancer patients. 2021. Proc. Natl. Acad. Sci. U. S. A. doi: 10.1073/pnas.2012228118
Technologies Developed
MetaboCore; single-cell on-chip metabolic cytometry (OMC)

Contact Dr. Wei Wei
Associate Professor
ISB