Project Summary
My Digital Gut – A Platform for Personalized Nutrition
The Gibbons Lab is developing an online dashboard called My Digital Gut to predict the impact of personalized dietary changes before people make them. The program assesses someone’s current diet (no food diary needed) and guides individuals towards personally optimal nutrition. Supported by the ISB Foundation Board, My Digital Gut is in the testing phase seeking to eventually inform personalized clinical care.
Executive Summary
ISB researchers are building an online dashboard to give clinicians and individuals the ability to optimize health with personalized dietary recommendations and other interventions, tailored to the composition of a gut microbiome. The dashboard, called My Digital Gut, will be powered by computational models designed to predict interventions that optimize the composition of the microbiome and its associated metabolites. My Digital Gut will be deployed in clinical trials, testing precision probiotic and dietary interventions, and has the potential to be used routinely by individuals to create actionable, personalized recommendations to improve their health.
Project At-A-Glance
- Funded by: Philanthropic donations and an Amazon Imagine Grant
- Led by Sean Gibbons, PhD
- Key Collaborators
- Andrew Magis, PhD, Nick Quinn-Bohmann, PhD, ISB
- Lance Anderson
My Digital Gut: Precision medicine to improve health by optimizing the gut microbiome
The human gut microbiome is vast in its complexity and variety. The gut is home to trillions of bacterial cells in hundreds of different species. These microbes help break down nutrients in our food, yielding tens of thousands of small molecules (metabolites) that we absorb into our body.
The microbiome is known to play a key role in human health. A properly balanced gut microbiome helps support immunity and shield individuals from disease. The gut microbiota has been shown to influence susceptibility to a host of chronic conditions, like type 2 diabetes, inflammatory bowel disease, and cardiovascular disease. Moreover, researchers at ISB and other institutions have demonstrated links between the microbiome and weight loss1 and healthy aging.2 Dr. Sean Gibbons and his colleagues have even shown that the microbiota influences how individuals respond to statins, cholesterol-lowering drugs.3
The gut microbiota shows enormous variability across humans. No two individuals have the same types and amounts of microbes in their gut. As a result, people vary widely in their responses to dietary, pharmaceutical, and lifestyle interventions.
Researchers like Gibbons are beginning to understand how interventions like foods, probiotics, and supplements may affect the microbiome and health – often through shifts in metabolism. A key goal is to tailor these interventions to an individual’s microbiome, to shield them from disease, reduce the incidence of unwanted side effects, and optimize their health. Toward this aim, Gibbons and his colleagues are generating computational models that incorporate personalized information on diet, microbiome composition and metabolism, to generate predictions of individual-specific outcomes of various interventions.
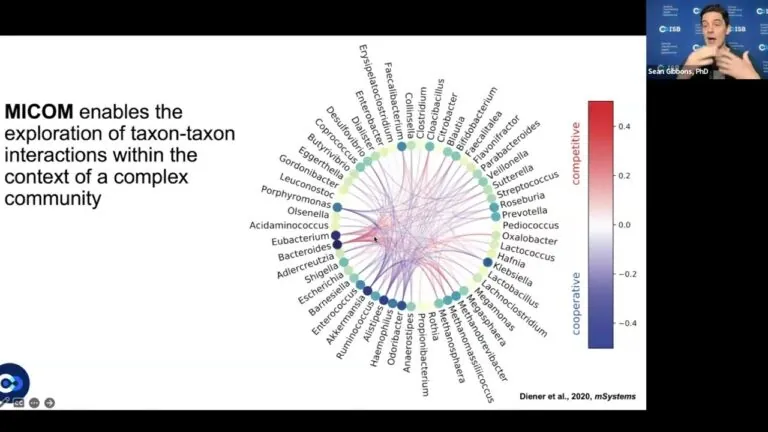
In 2020, Gibbons and his colleagues published one such model, dubbed MICOM (a portmanteau of Microbial Community).4 MICOM inputs nutritional information based on a standardized diet and data on an individual’s gut microbiome composition, derived from a stool sample. MICOM is widely used by the research community to explore interactions between microbes in the gut, dietary intake and ecosystem functions. ISB researchers are now building on MICOM to generate a next-generation model called CyberGut, which integrates host metabolism, and they are building another tool called MEDI (metagenomic estimation of dietary intake) that can be used to automatically assess dietary intake from stool sequencing data.5 Together, this ecosystem of tools will empower researchers to build host-microbe-diet models of the human gut that provide mechanistic insight into how the system works and predictions that may be leveraged to improve human health through precision nutrition.
MICOM, CyberGut, and MEDI are designed for use by researchers with expertise in computational biology. So, Gibbons and his team are now focused on building a user-friendly interface to extend their models to real-world applications, called My Digital Gut. My Digital Gut will be an online dashboard designed for use by physicians and their patients, linked up to the researchers’ evolving models.
Some of the first use-cases for My Digital Gut will be to support clinical trials. Gibbons and his colleagues are planning a trial to test how various supplements and probiotics may change patient responses to statins. The researchers are also discussing another trial to test the effects of precision dietary interventions for improving insulin sensitivity in healthy individuals.
In addition to supporting research, My Digital Gut also has the potential to become a personalized health tool, enabling individuals to optimize their health through targeted alterations to their microbiome. Gibbons envisions that after people sign up for My Digital Gut, they would receive a kit in the mail to provide a stool sample for processing. The data would be uploaded to the tool with user-friendly graphics showing their “digital twin,” an avatar that contains personalized information on the microbiome and diet. People could use the interface to digitally test the effects of various interventions, such as new types of diets (Mediterranean vs. Atkins, for example). Readouts might include resulting changes in key metabolites, such as the anti-inflammatory molecule butyrate, with information about their predicted effects on health. People might also have the option to consent to provide their de-identified data to researchers, who could use it to further hone and improve their models.
As researchers learn more about how various interventions can influence microbiome composition and health, the precision and accuracy of tools like My Digital Gut will increase. In the long term, the approach has the potential to radically change how disease is prevented and treated.
Citations
- Diener C, Qin S, Shou Y, Patwardhan S, Tang L, Lovejoy JC, Magis AT, Price ND, Hood L, Gibbons SM. Baseline Gut Metagenomic Functional Gene Signature Associated with Variable Weight Loss Responses following a Healthy Lifestyle Intervention in Humans. mSystems 2021 doi: 10.1128/mSystems.00964-21
- Wilmanski T, Diener C, Rappaport N, Patwardhan S, Wiedrick J, Lapidus J, Earls JC, Zimmer A, Glusman G, Robinson M, Yurkovich JT, Kado DM, Cauley JA, Zmuda J, Lane NE, Magis AT, Lovejoy JC, Hood L, Gibbons SM, Orwoll ES, Price ND. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab. 2021 doi: 10.1038/s42255-021-00348-0
- Wilmanski T, Kornilov SA, Diener C, Conomos MP, Lovejoy JC, Sebastiania P, Orwoll ES, Hood L, Price ND, Rappaport N, Magis AT, Gibbons SM. Heterogeneity in statin responses explained by variation in the human gut microbiome. Med 2022 doi: 0.1016/j.medj.2022.04.007
- Diener C, Gibbons SM, Resendis-Antonio O. MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota. mSystems 2020 doi: 10.1128/msystems.00606-19 \
- Diener C, Gibbons SM. Metagenomic estimation of dietary intake from human stool. bioRxiv 2024 doi: 10.1101/2024.02.02.578701v1
- Quinn-Bohmann, N, Wilmanski, T, Sarmiento, KR et al. Microbial community-scale metabolic modelling predicts personalized short-chain fatty acid production profiles in the human gut. Nat Microbiol. 2024 doi: 10.1038/s41564-024-01728-4
Technologies Developed
My Digital Gut, an online dashboard to predict the impact of personalized dietary changes and other interventions designed to affect the gut microbiome and associated metabolites.
MICOM (microbial community), a computational model that can predict tailored interventions to optimize the composition of an individual’s microbiome and its associated metabolites, thereby potentially improving health.
CyberGut, which extends MICOM to include host metabolism. These microbial community-scale metabolic models (MCMMs) will be used to power My Digital Gut.
MEDI (metagenomic estimation of dietary intake), a method to estimate dietary intake from the analysis of the DNA of consumed food items in stool.
How You Can Help
Help enable a new class of life-changing precision interventions for improving your gut health.
ISB Associate Professor Dr. Sean Gibbons is creating an online dashboard called My Digital Gut that will give anyone the ability to predict the impact of nutritional changes before making them. By supporting ISB, you can help us leverage our growing knowledge of the gut microbiome to make nutrition and healthcare personalized, predictive and preventive.
Thank You To Our Donors
We are grateful to the generous donors who have already supported My Digital Gut, believing in our vision to revolutionize personalized nutrition and healthcare through cutting-edge gut microbiome science. Your support made possible the development of a groundbreaking platform that will enable individuals and healthcare providers to make precision interventions promoting longer, healthier lives.