
Ultrafast Comparison of Personal Genomes
Genome sequences contain information with immense possibilities for research and personalized medical care, but their size, complexity and diversity make comparing sequences error-prone and slow. ISB researchers have created a method for summarizing a personal genome as a “fingerprint.”

Progress & Opportunities for Metabolic Reconstruction
In this study, we looked for evidence of such convergence through comparative analysis of 12 genome-scale yeast models.

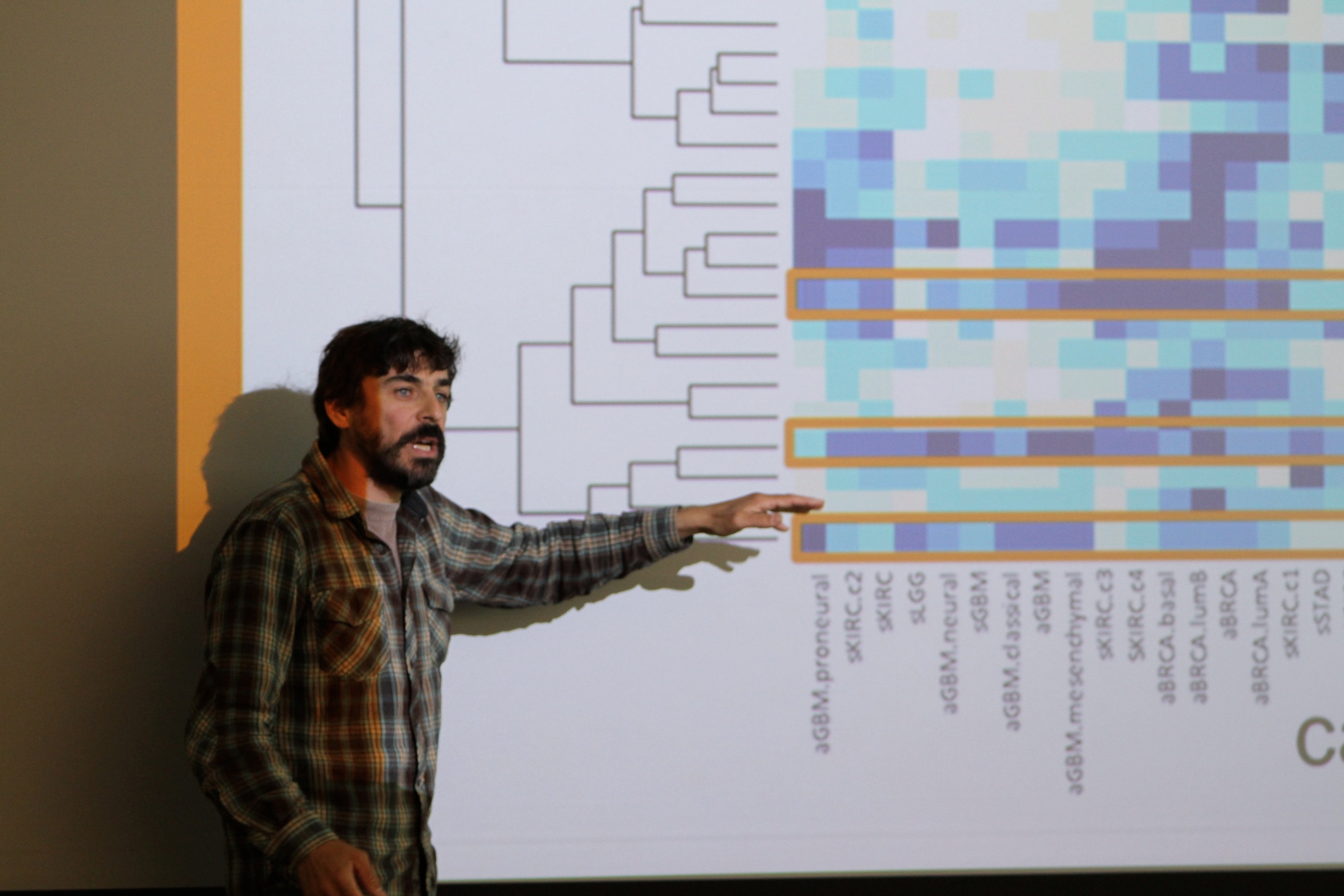
Prostate Cancer Study Identifies Numerous Subtypes of the Disease
Prostate cancer is the second most common cancer in men worldwide. The clinical behavior of prostate cancer is variable with some men exhibiting indolent prostate cancer which can be monitored over time while other men develop aggressive prostate cancer which can lead to metastasis and death.

Cancer Genomics Cloud
The ISB Cancer Genomics Cloud:
Leveraging Google Cloud Platform for TCGA Analysis
The ISB Cancer Genomics Cloud (ISB-CGC) is one of three pilot projects funded by the National Cancer Institute with the goal of democratizing access to the TCGA data by substantially lowering the barriers to accessing and computing over this rich dataset. The ISB-CGC is a cloud-based platform that will serve as a large-scale data repository for TCGA data, while also providing the computational infrastructure and interactive exploratory tools necessary to carry out cancer genomics research at unprecedented scales. The ISB-CGC will also facilitate collaborative research by allowing scientists to share data, analyses, and insights in a cloud environment.
Most Powerful Tool for Reconstructing a Gene Network
Nearly a decade ago, ISB’s Baliga Lab published a landmark paper describing cMonkey, an innovative method to accurately map gene networks within any organism from microbes to humans.
Two new papers describe the benchmark results of cMonkey and also the release of cMonkey2, which performs with higher accuracy.
Using this approach, genetic and molecular data generated from any organism, be it a bacterium or a birch tree, can be explored and analyzed from a network perspective.

Network Representations of Immune System Complexity
The mammalian immune system is a dynamic multiscale system composed of a hierarchically organized set of molecular, cellular, and organismal networks that act in concert to promote effective host defense. These networks range from those involving gene regulatory and protein–protein interactions underlying intracellular signaling pathways and single-cell responses to increasingly complex networks of in vivo cellular interaction,

Pushing the Molecular Switches of Tuberculosis Into Overdrive
Mycobacterium tuberculosis (MTB) infects more than 1.5 billion people worldwide partly due to its ability to sense and adapt to the broad range of hostile environments that exist within hosts.
To study how MTB controls its responses at a molecular level, ISB researchers and their collaborators at Seattle Biomed perturbed almost all MTB transcription factor regulators and identified the affected genes.
This comprehensive map of molecular switches in MTB provides a basis for understanding control mechanisms and can inform future efforts to rewire MTB responses for more favorable disease outcomes.

How Physics and Thermodynamics Help Assess DNA Defects in Cancer
‘Big data’ cancer research has revealed a new spectrum of genetic mutations across tumors that need understanding.
Existing methods for analyzing DNA defects in cancer are blind to how those mutations actually behave.
ISB scientists developed a new approach using physics- and structure-based modeling to systematically assess the spectrum of mutations that arise in several gene regulatory proteins in cancer.

New Details on Thyroid Cancer May Lead to More Precise Therapies
Papillary thyroid cancer represents 80 percent of all thyroid cancer cases.
Integrative analysis resulted in the detection of significant molecular alterations not previously reported in the disease.
ISB researchers identified microRNAs which may lead to more precise therapy.
BIOCELLION: New Supercomputer Software Framework Models Biological Systems at Unprecedented Scales
Computer simulation is a promising way to model multicellular biological systems to help understand complexity underlying health and disease.
Biocellion is a high-performance computing (HPC) framework that enables the simulation of billions of cells across multiple scales.
Biocellion facilitates researchers without HPC expertise to easily build and simulate large models.

Identifying Four New Subtypes of Gastric Cancer That May Lead to New Targeted Treatments
Gastric cancer has a high mortality rate, but current classification systems haven’t been effective in helping to identify subtypes relevant for treatment of the disease.
TCGA researchers have integrated molecular data from 295 stomach tumors and have discovered four subtypes of gastric cancer.
Stratification of patients into these four subtypes paves the way for the development of new personalized therapies.

Now researchers can explore genomic data across space and time
Understanding systems from a multiscale perspective gives us a more detailed and holistic view of how features or functions from each scale connect and interact in a given system.
The challenge is integrating the different types of information that come from each scale in an efficient way that yields the most insight.
ISB developed a new tool to make it easier for researchers to to integrate, analyze and visualize human genome data at multiple resolutions.
Powerful New Program Integrates Multiple Tools Into a Flexible Interface
Price Lab researchers worked with collaborators at the University of Illinois to create an easy-to-use software toolkit for comparing microbial genomes.
Tools can be used to find orthologs, correct missing/inaccurate gene annotations, analyze gene gain and loss patterns, and build draft metabolic networks from reference networks.
Software offers predictive and analysis capabilities in a flexible way, making it easy to build customized analysis pipelines.

ISB Researchers Develop New Method to Study and Predict Traits of Cells
ISB researchers used the systems approach to develop a new way to integrate data from different classes of networks to better understand how cells function.
The method is a software program called GEMINI and it’s the first of its kind to integrate data from metabolic networks to refine transcriptional regulatory networks.
GEMINI has higher success rate than existing technologies.

A ‘Sound Check’ for Digital Transcriptomes
Looking for biomarkers in different types of tissues requires comparing massive amounts of gene expression data.
In order to compare ‘digital transcriptome’ data, they have to be normalized or adjusted to a common standard of measurement.
ISB researchers developed new algorithmic methods that outperform existing methods for normalizing gene expression data from different samples.

How ISB’s Systems Approach Finds New Biological Insight from Existing Big-Data Sets
There exist copious amounts of public research data that can reveal new biological information if they are integrated and analyzed.
One of ISB’s specialties is the ability to apply systems approaches to develop methods to integrate and analyze data.
In the latest publication, ISB demonstrates an open-access computational strategy that can help any researcher capitalize on large data sets.



