How Microbes Learn to Predict the Future
Like plants and animals, even microbes can anticipate and prepare in advance for future changes in their environment
Similar to how Pavlov trained a dog to anticipate food when it heard a bell, ISB researchers trained yeast to anticipate a lethal toxin when it sensed caffeine
The study revealed how in a very short period of time yeast can evolve to ‘learn’ and ‘predict’ new patterns in their environment
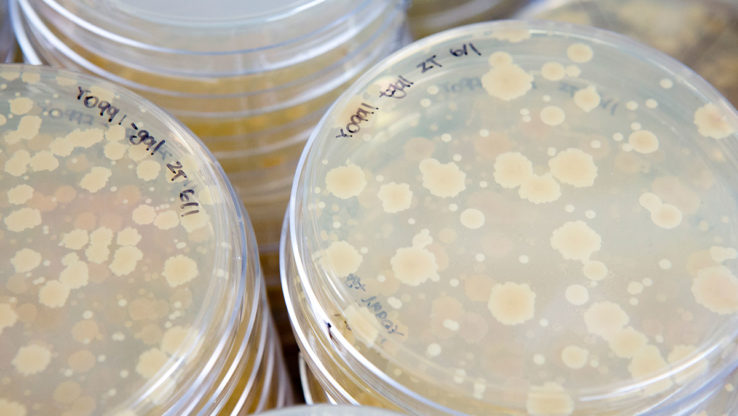
Introduce One-cell Doubling Evaluation of Living Arrays of Yeast! ODELAY!
Genes, Genomes, Genetics, scientists at Institute for Systems Biology introduce ODELAY, a powerful automated and scalable growth analysis platform that uses time-lapse microscopy to photograph individual yeast cells growing into colonies.
Predicting Cell Fate Decisions Using Single-Cell Analysis and the Theory of Tipping Points
ISB researchers developed a new theory to exploit burgeoning single-cell molecular profiling measurements to make predictions of future cell behaviors.
New Tool Uses 3-D Protein-DNA Structures to Predict Locations of Genetic ‘On-Off’ Switches
Novel systems approach uses high-resolution structures of protein-DNA complexes to predict where transcription factors (genetic switches) bind and regulate the genome.
This approach can help researchers better understand and predict binding sites for non-model organisms or ‘exotic’ species.
Having such insight and predictive capabilities is critical for reverse- and forward-engineering organisms that could be pivotal for new green biotechnologies.
Unpredictable Environments Require Predictably Fast Responses
ISB researchers discover a novel mechanism used by cells to rapidly turn “on” or “off” genes in order to change survival strategies in response to environmental events.
The “switch” was discovered in Halobacterium salinarium and found to be conserved in diverse life forms.
Research shows that microbes may use specialized switches depending on the type of environmental challenge they encounter.
Tiniest Malfunctions in a Cell Can Cause Devastating Diseases
ISB researchers are studying peroxisomes, which are cellular organelles that are linked to a rare syndrome that causes progressive organ complications and infant mortality.
Peroxisomes have a role in metabolizing and breaking down cellular waste.
Because peroxisomes easily change shape and function according to a cell’s needs, a systems approach is necessary to help decipher that complexity.

ISB Researchers Develop New Method to Study and Predict Traits of Cells
ISB researchers used the systems approach to develop a new way to integrate data from different classes of networks to better understand how cells function.
The method is a software program called GEMINI and it’s the first of its kind to integrate data from metabolic networks to refine transcriptional regulatory networks.
GEMINI has higher success rate than existing technologies.

How ISB’s Systems Approach Finds New Biological Insight from Existing Big-Data Sets
There exist copious amounts of public research data that can reveal new biological information if they are integrated and analyzed.
One of ISB’s specialties is the ability to apply systems approaches to develop methods to integrate and analyze data.
In the latest publication, ISB demonstrates an open-access computational strategy that can help any researcher capitalize on large data sets.
Evaluating Effects of Variable Single-Cell Protein Expression on Metabolism
Stochastic gene expression can lead to phenotypic differences among cells even in isogenic populations growing under macroscopically identical conditions. Here, we apply flux balance analysis in investigating the effects of single-cell proteomics data on the metabolic behavior of an in silico Escherichia coli population. We use the latest metabolic reconstruction integrated with transcriptional regulatory data to model realistic cells growing in a glucose minimal medium under aerobic conditions.

New Open-Access Software Helps Predict Cellular Actions Tied to Diseases, Drug Targets
All living things are made of cells that contain DNA, which help determine their physical characteristics. In addition to this encoded genetic information, organisms are also defined by the way they decode information from interactions with their environments. Signals from the environment are interpreted by cells through a series of steps that turn proteins on or off, making up what is often referred to as the “signaling network.”
Role of Key Nuclear Pore Proteins on the Assembly of Chromatin
The three-dimensional architecture of a cell’s nucleus, which contains its genetic information, is critical to determining which of a cell’s genes are actually expressed. Using systems and cell biology techniques, researchers have identified how nuclear pore complexes—collections of proteins studding the nuclear membrane— serve as key elements in defining nuclear architecture and controlling which genes get expressed. This new insight into the behavior of nuclear pore complex proteins frequently targeted by infectious diseases is published in the Feb. 28 issue of the journal Cell, which is among the top-five most prestigious journals in the country.



